Tutorial on binning, PDFs, CDFs, 1-CDFs and more
Introduction
In this course, we will need to plot various empirical probability distributions. As we deal with data, whose sparsity, and order of magnitudes may vary a lot, we have provided this tutorial to help you in producing appropriate visualizations of the data. But first, some mathematical background:
Discrete variables: Probability mass functions (PMF)
Let us assume we have some random variable \(V\) that can have only discrete values. Then the function describing the probabilities for different outcomes is a probability mass function (PMF) \(P(v)\). If we assume, for simplicity, that our random variable \(V\) can only take integer values, the probabilities for obtaining different values of \(V\) can be written as:
Probability of V being v (\(P(V=v)\)) is simply written as \(P(v)\)
Probability of V being some value between \(x\) and \(y\) (with \(x\) and \(y\) included) equals
\[\sum_{v=x}^y P(v)\]
The probability mass function is also normalized to one:
\[\sum_{i=-\infty}^\infty P(i) = 1 \]
Continuous variables: Probability density function (PDF)
The counterpart of a PMF for a continuous random variable \(V\) is its probability density function (PDF), denoted also typically by \(P(v)\). For a probability density function \(P(v)\) it holds:
Probability of observing a value between \(x\) and \(y\) \((>x)\) equals
\[\int_x^y P(v) dv\]
The distribution is normalized to one:
\[\int_{-\infty}^{\infty} P(v) dv = 1\]
Example of a PDF denoted by f(x):
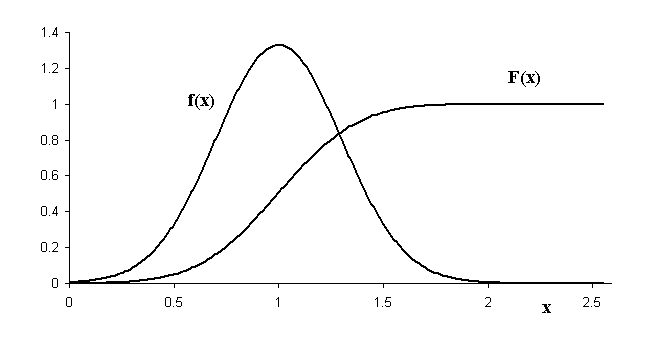 (Figure from: http://physics.mercer.edu/hpage/CSP/pdf-cpf.gif)
(Figure from: http://physics.mercer.edu/hpage/CSP/pdf-cpf.gif)
(\(F(x)\) denotes the cumulative density function, more of that later)
Note that PDFs can have values greater than 1!
From now on in this tutorial, we mostly assume that we work with continuous random variables
In other words, we assume that the elements in our data are real numbers that arise from a distribution that is continously distributed, and described by a probability density function (PDF). In practice, the same methods can (usually) be used also when we are dealing with discretely distributed data, such as node degrees in a network.
Computing and plotting empirical PDFs:
Let us start by presenting data that are either (i) "narrowly" distributed, or (ii) have a fat-tail using standard matplotlib plotting settings:
Solution: logarithmic bins:
Use logarithmic bins instead of linear!
Now we can see something in the tail as well! What is the slope of the PDF of 1/x, with \(x~Uniform(0,1)\) in the log-log scale? (For the interested: can you derive the analytical form of the PDF and calculate the slope using pen and paper?)
For intepreting log-log PDF plots, one just needs to know how different "standard" distributions look like (on a log-log scale).
Don't want to bin? Use CDF(x) and 1-CDF(x)!
Representing PDFs using binning strategies can be difficult due to limited samples of data etc.. To neglect the need for binning, one can use the cumulative distribution function (CDF) and the complementary cumulative distribution function (1-CDF).
Cumulative density function (CDF(x)):
The standard mathematical definitions:
Discrete variable: \(CDF(x) = \sum_{i \leq x} P(x)\)
Continuous variable: \(CDF(x) = \int_{-\infty}^{x} P(x') dx\)
Complementary cumulative density function (cCDF(x), 1-CDF(x))
- In this course, the complementary cumulative distributions are often more practical.
- 1-CDF is especially useful when dealing with fat-tailed distribution as they enable one to zoom in to the tail of the distribution.
In this course we will use the following definitions for "1-CDF(x)":
Discrete variable: \(1-CDF(x) = \sum_{i \geq x} P(x)\)
Continuous variable: \(1-CDF(x) = \int_{x}^{\infty} P(x') dx\)
Note:
The standard definition of the discrete "1-CDF(x)" does not take into count in the values that equal x. Thus our above "1-CDF(x)" + "CDF(x)" does not exactly equal one. The reason why we take up our nonstandard 1-CDF(x) definition is that it enables us to work more practically with real data. Especially, were we to plot the 1-CDF(x) on a logarithmic y-axis, the largest observed data point that we have would not be visible with the standard 1-CDF(x) definition. (Why?)
So given some data vector \(d\) data, we present the empirical 1-CDFs as: \[1-CDF(x) = \frac{\text{number of elements in $d$ that are $\geq x$}}{\text{number of elements in $d$}}\]
Plotting 2D-distributions
What is the dependence between values of vectors X and Y?
First approach: simple scatter plot :
Can we spot the slight trend?
Compute bin-averages! (with binned_statistic):
More sophisticated approaches for presenting 2D-distributions include e.g. heatmaps, which can be produced using using binned_statistic2d and pcolor. More of those later in the course!
 (Figure from: http://physics.mercer.edu/hpage/CSP/pdf-cpf.gif)
(Figure from: http://physics.mercer.edu/hpage/CSP/pdf-cpf.gif)